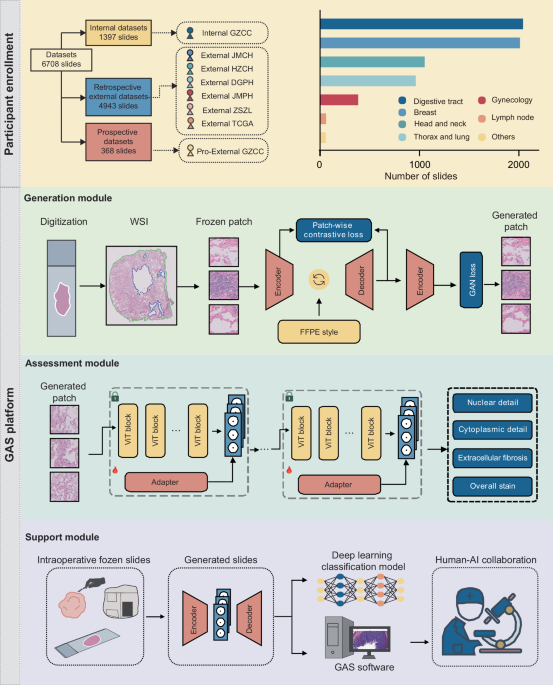
Patient cohorts
This multicenter study included retrospective development and validation of the GAS platform, followed by a prospective validation to assess its generalizability. The Generation module was trained using retrospectively collected data from the Internal GZCC. External validation involved six independent cohorts from JMCH, HZCH, DGPH, JMPH, ZSZL, and TCGA. A prospective cohort was enrolled at Pro-External GZCC. All included patients had intraoperative diagnoses and matched post-operative FFPE-confirmed diagnoses. Frozen and FFPE slides were derived from the same surgical specimens, though exact pairing was not required due to the unpaired GAN architecture. Exclusion criteria included patients with incomplete clinical data and slides with extensive tissue folds, large fractures, or severe out-of-focus regions. The study was approved by the ethics committee of Sun Yat-sen University Cancer Center (No. SL-B2023-416-02), with informed consent waived for retrospective data and obtained for the prospective cohort.
Data digitalization
For each case, all available tumor slides were scanned at ×40 magnification (0.25 μm/pixel) for further analysis. Slides from the Internal GZCC, External HZCH, External DGPH, and Pro-External GZCC cohorts were scanned with a PHILIPS Ultra-Fast Scanner (Philips Electronics N.V., Amsterdam, Netherlands) and saved in iSyntax format. Meanwhile, slides from the External JMCH and External JMPH datasets were scanned using the Aperio AT2 scanner (Leica Biosystems, Wetzlar, Germany) and stored in SVS format.
Data preprocessing
For each digitized slide, CLAM’s preprocessing24 method was used to segment the tissue regions in the WSIs. Based on this segmentation mask, the tissue regions were partitioned into non-overlapping 512 × 512 patches at 20× magnification, with coordinates retained for the final reconstruction of the entire slide. Due to imperfections in the segmentation, partial background regions such as text or partial blank areas were often included. It was observed that background patches erroneously included exhibit distinctly different histogram distributions compared to tissue patches (Supplementary Fig. 8). To eliminate these background patches, all patches were converted to grayscale, and the mean and variance of their grayscale images were computed. Background patches were then filtered based on thresholds applied to these statistics. In addition, within the remaining patches, there were areas blurred due to lens defocus during slide scanning, which were identified and removed using the Laplacian method25.
Generation module
Model overview: To synthesize FFPE-like images from frozen sections, we developed the Generation module of the GAS platform, comprising an encoder, a style neck, and a decoder. The encoder downsampled inputs to expand the receptive field and reduce computational burden. The style neck enabled domain translation, and the decoder reconstructed the output, aided by skip connections to retain fine details lost during downsampling. Training optimized a weighted sum of adversarial26 and PatchNCE losses27. The adversarial loss drove global realism, while PatchNCE preserved local structural fidelity by maximizing mutual information between input and output patches.
Style neck: The style neck was composed of stacked residual blocks28. Crucially, the normalization layers within the residual blocks were Adaptive Instance Normalization (AdaIN) layers29, which accomplished the style transformation of the input images. The AdaIN layer altered the style of the input image through adaptive affine transformations (Eq. (1)):
$${AdaIN}\left(x,y\right)=\sigma \left(y\right)\left(\frac{x-\mu \left(x\right)}{\sigma \left(x\right)}\right)+\mu \left(y\right)$$
(1)
where x represented the input image and y represents the target style image. μ(·) and σ(·) denoted the mean and standard deviation, respectively. The original AdaIN layer required multiple computations of the affine parameters (the mean and standard deviation) from the style input. We proposed that the affine parameters could be directly predicted from text descriptions of the FFPE style, without the need for additional input images of the FFPE style and repeated calculations. There, we employed the text encoder of QuiltNet30 to text encoder encode the text representing the FFPE style, which was “formalin-fixed paraffin-embedded tissues”. The encoded text was then fed into a multilayer perceptron (MLP) to predict the affine parameters. Through this design, our generative model efficiently leverages pathology-specific knowledge embedded in the pretrained network to encode FFPE-style representations. These representations guide the image synthesis process, enabling the model to generate high-quality FFPE-like images from frozen section inputs.
Training configurations: Our model was optimized using the Adam31 optimizer with a learning rate set to 0.0002. It underwent training with a batch size of 1 over a maximum of 200 epochs. To ensure a fair comparison, all baseline models were trained under consistent settings. The generative model was trained with flip-equivariance augmentation, where the input image to the generator was horizontally flipped, and the output features were flipped back before computing the PatchNCE loss.
Differences from our method: Unlike prior approaches, our method is tailored for intraoperative scenarios, where rapid inference is essential. To this end, we use a lightweight encoder–decoder architecture with fewer layers and incorporate skip connections to mitigate feature loss during downsampling. Rather than relying on computationally intensive attention modules, we leverage domain knowledge embedded in a pathology-pretrained model to generate textual descriptions of FFPE characteristics. These are integrated into the generator via AdaIN, enabling fast and high-fidelity generation of FFPE-style images.
Reader study: To assess the fidelity of the generated images compared to real FFPE images, a reader study was conducted. Three pathologists reviewed 200 image patches (100 generated and 100 real FFPE) and classified them as either generated or real, and reviewed 200 image patches (100 generated and 100 frozen) and classified them as either generated or frozen.These image patches were randomly sampled from all test sets, and the sampling process was unbiased. Regarding tissue region selection, the preprocessing pipeline was consistent across datasets.
Assessment module
With the application of foundation models in computational pathology15,32,33,34, the ability of these models to extract general features from pathological images has significantly improved. We developed the Assessment module using pathological image quality control models derived from the foundation model. Then, the adapter architecture was introduced to enhance the model’s performance on the task of pathological image quality assessment. During training, the parameters of the foundation model were kept frozen, only the adapter and the projector used for prediction were trained. The foundation model consisted of 24 ViT blocks35. The adapter followed an MLP architecture. To retain the original knowledge encoded by the foundation model, a residual structure was introduced. Specifically, the output of the ViT block was fed into the adapter, and the adapter’s output was aggregated with the original features using a fixed parameter γ (Eq. (2)):
$${{\mathcal{F}}}_{l}^{* }=\gamma {A}_{l}{({F}_{l})}^{T}+(1-\gamma ){F}_{l}$$
(2)
Where Al denoted the adapter at layer l, Fl represented the output of the ViT block at layer l, and \({{\mathcal{F}}}_{l}^{* }\) indicated the output feature, which served as the input to the subsequent ViT block, and γ was set to 0.8 in this experiment36,37.
Compared to Low-Rank Adaptation (LoRA)38, a widely used parameter-efficient fine-tuning method that modifies model weights through low-rank matrix updates, the adapter approach adjusts features via external modules36,39,40. It combines original and adapted features using a scaling factor γ, allowing finer control over the degree of adaptation. As the foundation model, UNI21, was pretrained on over 100 million pathology images using the MoCoV341 framework and already captures extensive domain knowledge, the adapter strategy preserves core representations while introducing task-specific features for quality control, thereby improving overall performance.
The quality control models were trained using a batch size of 16, with training lasting up to 50 epochs. Optimization employed the Adam optimizer with an L2 weight decay set at 5e−5, along with a learning rate of 1e−5. Segmentation and patching of WSIs were performed on Intel(R) Xeon(R) Gold 6240 Central Processing Units (CPUs), and the models were trained on 4 NVIDIA GeForce RTX 2080 Ti Graphics Processing Units (GPUs).
To verify whether the quality control model attended to relevant features, we applied Grad-CAM. A forward pass produced the final convolutional feature map and the pre-softmax prediction score. Gradients of the score with respect to the feature map were then computed via backpropagation. Channel-wise importance weights were calculated, aggregated through a weighted sum, and passed through ReLU to generate the final localization map.
Deep-learning classification of support module
To evaluate whether the synthetic FFPE-like images generated by GAS provide greater utility in downstream classification tasks compared to frozen images, we designed a series of deep-learning classification tasks. These included: margin assessment, sentinel lymph node metastasis prediction in breast cancer, classification of benign versus malignant thyroid lesions, lung adenocarcinoma versus lung squamous cell carcinoma, benign versus malignant breast lesions, and breast carcinoma in situ versus invasive breast cancer.
For the task of distinguishing lung adenocarcinoma from lung squamous cell carcinoma, cases from TCGA were used to develop the model. For the classification of benign versus malignant thyroid lesions, benign versus malignant breast lesions, and carcinoma in situ versus invasive breast cancer, FFPE data were used for model development. In contrast, for margin assessment and sentinel lymph node metastasis tasks, diagnostic models were developed using either original frozen images or GAS-generated images only, without incorporating any FFPE data. The classification results obtained using GAS-generated images were compared with those using frozen images as inputs. The prediction process using the generated images was as follows: tissue regions in the frozen images were segmented and divided into 512 × 512 patches. Subsequently, synthetic FFPE-like patches were generated from frozen patches, and their features were extracted. Finally, the patch-level features were aggregated into a slide-level feature for prediction. The feature aggregation follows the attention-based pooling function introduced in CLAM (Eq. (3)):
$${a}_{i,k}=\frac{{e}^{\{{W}_{a,i}(\tan \,{\rm{h}}\,{V}_{a}{{\bf{h}}}_{k})\odot simg({U}_{a}{{\bf{h}}}_{k})\}}}{{\sum }_{j=1}^{K}{e}^{\{{W}_{a,i}(\tan \,{\rm{h}}\,{V}_{a}{{\bf{h}}}_{j})\odot simg({U}_{a}{{\bf{h}}}_{j})\}}}$$
(3)
$${{\boldsymbol{f}}}_{{slide},i}=\mathop{\sum }\limits_{k=1}^{K}{a}_{i,k}{{\boldsymbol{h}}}_{k}$$
Here, fslide,i denotes the aggregated feature representation for class i, ai,k is the attention score for patch k, and hk is the patch-level feature. Va and Ua are learnable fully connected layers, and Wa,i epresents one of N parallel attention branches.
GAS software of support module
A prospective assessment using a sub-cohort from the Pro-External GZCC cohort was conducted to validate the clinical utility of GAS in intraoperative diagnostic workflows. We developed a human–AI collaboration software to facilitate the process. Once the frozen sections were prepared, they were scanned to create digital slides. After uploading these slides to the GAS software, pathologists can browse the frozen sections online. If any areas appear unclear, the pathologists can click on the corresponding regions, and the software will convert these unclear areas into high-resolution FFPE-like images.
Quantification and statistical analysis
FID was used to measure image similarity for the generative models. Additionally, the quality control model evaluated the generative models using scoring as another metric. For the quality control model, accuracy and AUROC served as the primary evaluation metrics. The external test sets from different centers, allowed for a more comprehensive assessment of the generalization properties of GAS. A P value less than 0.05 was considered statistically significant. Statistical significance was evaluated using t test and Wilcoxon signed-rank test. Data preprocessing and model development were conducted using Python (version 3.8.0) and the deep-learning platform PyTorch (version 2.3.0).








Leave feedback about this